Arrays and Matrices in R
Arrays are declared in R using the dim() or array() functions.
For example:
> mycarsale.array<-1:30
> mycarsale.array
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
[17] 17 18 19 20 21 22 23 24 25 26 27 28 29 30
The above command allocates a vector of 1 to 30 to the variable array called mycarsale.array
To make it as a matrix, we can use dim() function as given below
> dim(mycarsale.array)<-c(3,5,2)
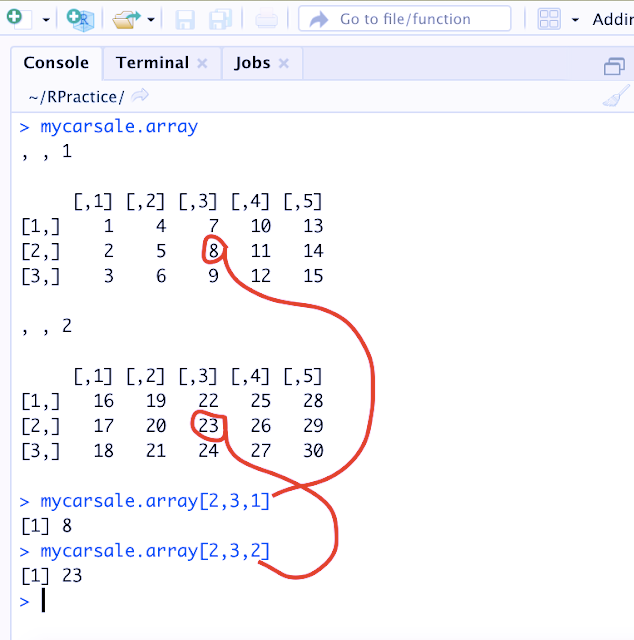
> mycarsale.array
, , 1
[,1] [,2] [,3] [,4] [,5]
[1,] 1 4 7 10 13
[2,] 2 5 8 11 14
[3,] 3 6 9 12 15
, , 2
[,1] [,2] [,3] [,4] [,5]
[1,] 16 19 22 25 28
[2,] 17 20 23 26 29
[3,] 18 21 24 27 30
In the above output 3 indicates the number of columns, 5 indicate the number of columns and 2 indicates the number of tables.
 |
| Array in R |
> mycarsale.array[2,3,1]
[1] 8
> mycarsale.array[2,3,2]
[1] 23
Comments
Post a Comment